Boutry Hofmann Norris the West Guide to Writing Pdf
Abstract
A locus at 19p13 is associated with breast cancer (BC) and ovarian cancer (OC) risk. Here we analyse 438 SNPs in this region in 46,451 BC and 15,438 OC cases, 15,252 BRCA1 mutation carriers and 73,444 controls and identify 13 candidate causal SNPs associated with serous OC (P=9.2 × 10−20), ER-negative BC (P=1.1 × 10−13), BRCA1-associated BC (P=7.7 × 10−16) and triple negative BC (P-diff=2 × 10−5). Genotype-gene expression associations are identified for candidate target genes ANKLE1 (P=2 × 10−3) and ABHD8 (P<2 × 10−3). Chromosome conformation capture identifies interactions between four candidate SNPs and ABHD8, and luciferase assays indicate six risk alleles increased transactivation of the ADHD8 promoter. Targeted deletion of a region containing risk SNP rs56069439 in a putative enhancer induces ANKLE1 downregulation; and mRNA stability assays indicate functional effects for an ANKLE1 3′-UTR SNP. Altogether, these data suggest that multiple SNPs at 19p13 regulate ABHD8 and perhaps ANKLE1 expression, and indicate common mechanisms underlying breast and ovarian cancer risk.
Introduction
Genome-wide association studies (GWAS) have identified more than 100 different genetic susceptibility regions for breast cancer (BC)1,2,3,4,5,6 and 20 regions for epithelial ovarian cancer (EOC)7,8,9,10,11,12,13. A few of these regions, and in some cases the same genetic variants, are associated with risks of both cancers (pleiotropy), suggesting there may be underlying functional mechanisms and biological pathways common to different cancers. The TERT-CLPTM1L locus (5p15) is one such example in which the same variants are associated with risks of oestrogen receptor (ER)-negative BC, BC in BRCA1 mutation carriers and serous invasive OC10.
Few studies have comprehensively described the functional mechanisms underlying common variant susceptibility loci10,14,15,16,17,18. More than 90% of risk alleles lie in non-protein-coding DNA and there is now unequivocal evidence that susceptibility regions are enriched for risk-associated single-nucleotide polymorphisms (SNPs) intersecting regulatory elements, such as transcriptional enhancers, predicted to control the expression of target genes in cis 19,20,21. Establishing causality for risk SNPs is very challenging; of the thousands of risk associations identified by GWAS, functional validation of causal variants using genome editing has only been experimentally performed for two SNPs, one for prostate cancer22 using the CAUSEL pipeline and the other for obesity23. Thus, there is a critical need to identify the causal risk SNP(s) and the overlapping regulatory element(s) and the target gene(s) regulated in an allele-specific manner.
Breast and high-grade serous OC share common genetic and non-genetic risk factors, with mutations in BRCA1 and BRCA2 the most significant risk factors for both cancers, suggesting similar biological mechanisms drive breast and OC development. A region on chromosome 19p13.1 has previously been associated with susceptibility to BC and OC in the general population, and to modify the risks of BRCA1-related BC and BRCA2-related OC9,24,25,26,27. Initial studies indicated that the association signal was centred around the SNP rs8170 located in the BRCA1-interacting gene BABAM1 (ref. 9), and subsequent studies have refined the subtype specific BC risks associated with these SNPs24,25,26,28.
In the current study, we hypothesized that the same functional mechanism underlies the 19p13.1 risk association in both BC and OC. To evaluate this hypothesis we performed genetic fine mapping in BC and OC patients and in BRCA1 mutation carriers, and performed a wide range of functional assays in breast and ovarian tissues and in vitro models to identify the likely causal alleles, and target regulatory elements and susceptibility gene(s). Our data indicate that multiple SNPs are involved in the regulation of ABHD8 and perhaps ANKLE1 at this locus.
Results
Genetic association analyses with breast and OC risks
A total of 438 SNPs spanning 420 kb at the chromosome 19p13 locus (nucleotides 17,130,000–17,550,000 (NCBI build 37)) were genotyped successfully in the following populations: 46,451 BC cases (of which 7,435 cases had ER-negative tumours) and 42,599 controls from the Breast Cancer Association Consortium (BCAC); 15,438 cases of EOC (of which 9,630 were of serous histology) and 30,845 controls from the Ovarian Cancer Association Consortium (OCAC); and 15,252 BRCA1 mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA; 7,797 with BC and 7,455 unaffected; Supplementary Table 1). Genotypes for variants identified through the 1,000 genomes project (minor allele frequency (MAF)>0.1%) were imputed for all participants of European ancestry. A total of 2,269 genotyped and imputed SNPs were analysed for their associations with ER-negative BC risk in the general population, 2,311 SNPs with BC/OC risk for BRCA1 mutation carriers, and 2,565 SNPs with risk of serous OC. Results for all SNPs associated with these phenotypes at P<10−4 are illustrated in Fig. 1 and Supplementary Fig. 1. Two perfectly correlated SNPs rs61494113 and rs67397200 located between the ANKLE1 and ABHD8 genes demonstrated the strongest association with BC risk among BRCA1 mutation carriers (χ 2-test P=7.8 × 10−16) and ER-negative BC in BCAC (χ 2-test P=1.3 × 10−13, P-meta-analysis=7.3 × 10−28). There was no association for ER-positive BC (χ 2-test P=0.21 for rs61494113). The strongest association with invasive and serous OC was for rs4808075 (correlated with rs61494113 with r 2=0.99) located in the BABAM1 gene (χ 2-test P=9.2 × 10−20). We observed no associations with risk of other histological subtypes of invasive OC (Supplementary Table 2). The correlations between the SNP exhibiting the strongest risk association (rs67397200) in the meta-analysis of BC risk for BRCA1 mutation carriers and ER-negative BC, with the previously reported risk-associated SNPs for breast, OC and BRCA1-associated BCs can be found in Supplementary Table 3.

Results for ER negative breast cancer from BCAC, for ovarian cancer from OCAC and for BRCA1 mutation carriers with breast cancer from CIMBA are shown. Also shown are the results of a meta-analysis for BRCA1 and general population ER negative breast cancer cases. The grey bars indicate the boundaries of the two association peaks, and the dotted horizontal line indicates the cutoff for genome-wide significance (χ 2-test P=5 × 10−8). Previously identified GWAS SNPs are indicated with italic font. Genes in the region are displayed beneath the association results.
Full size image
All SNPs with an association P value<0.001 with each phenotype were included in forward stepwise Cox regression models for risks of BRCA1 BC, and logistic regression models for ER-negative BC and serous OC. The most parsimonious models for ER-negative BC and serous OC each included one SNP, rs67397200 for ER-negative BC and rs4808075 for serous OC (referred to as Peak 1). The most parsimonious model in the analysis of BC risk for BRCA1 mutation carriers included two virtually uncorrelated SNPs (pairwise correlation r 2=0.018) rs61494113 (P value=4.4 × 10−16 in conditional regression analysis), and rs3786515 (Peak 2, conditional regression P value=9.6 × 10−5, pairwise correlation r 2=0.018; Fig. 1). No other SNP was retained in the model at the P value threshold of 0.0001.
Candidate causal variants
Peak 1 includes SNPs that encompass the BABAM1, ABHD8 and ANKLE1 gene and are associated with serous OC, ER-negative BC and BC risk for BRCA1 mutation carriers (Fig. 1 and Supplementary Fig. 1); Peak 2 includes SNPs located in the MYO9B gene associated only with BC risk in BRCA1 mutation carriers. SNPs in Peaks 1 and 2 are virtually uncorrelated.
To identify the strongest candidate causal SNPs, we computed likelihood ratios of each SNP relative to the SNP with the strongest association in each peak for risks of each phenotype. Due to the similarities in associations between ER-negative BC and BRCA1-associated BC in Peak 1, we computed the likelihood ratios on the basis of the meta-analysis results. Table 1 includes the SNPs that cannot be excluded at a likelihood ratio of >1:100 fold. In Peak 1, all but 12 SNPs can be excluded from being causal for ER-negative BC and BRCA1-associated BC. An additional SNP (rs10424198) cannot be excluded from being causal for serous OC. All 13 SNPs were highly correlated (r 2>0.95) and spanned a region of 19.4 kb. In Peak 2, the likelihood ratios of each SNP were calculated on the basis of the BRCA1 association analysis conditional on the top SNP rs61494113. All but seven SNPs correlated with rs3786515 (r 2>0.10) cannot be excluded from being the causal SNP for BRCA1-associated BC risk. With the exception of rs3786514 (pairwise r 2 with rs3786515=0.87) all other SNPs had r 2 with rs3786515 between 0.13 and 0.20.
Full size table
Associations for BRCA1 and BRCA2 mutation carriers
SNPs in Peak 1 were only associated with risk of ER-negative BC for BRCA1 mutation carriers and provided no evidence of association with ER-positive BC for BRCA1. SNPs in Peak 1 were also associated with OC risk for BRCA1 mutation carriers. SNPs in Peak 2 were also primarily associated with BRCA1-related ER-negative BC but there was no evidence of association with OC risk (Supplementary Table 4). SNPs in peak 1 were not associated with overall risk of BC in BRCA2 carriers (for example, rs67397200 HR for BC=1.00 (95% confidence interval (CI): 0.93–0.89)); however, SNP rs67397200 showed evidence of association with OC for BRCA2 mutation carriers (hazards ratio (HR)=1.18, 95% CI: 1.06–1.36, χ 2-test P=0.0056). SNPs in peak 2 did not show any evidence of association with breast or OC risk for BRCA2 mutation carriers.
Associations with risk among BC subtypes
None of the Peak 1 SNPs were associated with risk of ER-positive BC. When analyses were restricted to triple negative BC, the odds ratio (OR) estimates for SNPs in Peak 1 were larger than the corresponding OR estimates for ER-negative disease (Supplementary Table 4). There was no evidence of association with ER-negative and HER2-positive BC risk, with the association restricted only to triple-negative BC (test of difference between triple-negative versus ER-negative/HER2+, P-diff=2.2 × 10−5 for SNP rs61494113).
Analysis in Asian and African ancestry studies
None of the SNPs in the fine-mapping region were associated with ER-negative BC in samples of Asian ancestry after adjusting for multiple testing (P values≥0.0018). However, the risk alleles of the 13 candidate causal SNPs in Peak 1 are uncommon in the Asian population (MAF=0.0079–0.011); hence, the power to detect an association was limited and, due to the wide CIs for the estimated ORs for these SNPs, we cannot rule out that the minor allele of these SNPs in Asian subjects is associated with similar level of risk as in Europeans. In samples of African ancestry only rs4808616 (MAF=0.22) showed evidence of association with risk for overall BC or ER-negative disease (OR for BC=1.19, 95% CI:1.02–1.39, χ 2-test P=0.03; OR for ER-negative BC=1.59, 95% CI: 1.02–2.49, χ 2-test P=0.04).
Functional characterization of the 19p13.1 region
Functional characterization focused on the 13 candidate causal SNPs for ER-negative and BRCA1-associated BC and serous OC in Peak 1, based on the hypothesis that the functional mechanisms mediated by one or more of these SNPs were the same for these phenotypes.
Genotype-gene expression associations
We used expression quantitative trait locus (eQTL) analyses to evaluate associations between risk SNPs and the expression of genes in a 1 Mb region spanning rs4808075 in: 135 normal breast tissues29, 60 normal ovarian and fallopian tube epithelial cell cultures, 391 ER positive BCs30, 59 ER-negative BCs29 and 340 high-grade serous OCs30. We identified significant eQTL associations for ABHD8 expression (linear regression P value range 2 × 10−3–7 × 10−3) in normal breast tissues and between rs480816 and ABHD8 expression in OCs (linear regression P=3 × 10−5). In both instances the risk allele was associated with higher ABHD8 expression (Fig. 2a, Supplementary Data 1 and 2 and Supplementary Table 5). We examined whether risk SNPs were the top eQTL SNPs in this region. rs4808616 was the strongest predictor of ABHD8 expression in OCs. However, in normal breast tissues the top eQTL SNP for ABHD8 was rs11666308 (linear regression P=3.3 × 10−4), a marginally better predictor than rs4808616 (linear regression P=2.8 × 10−3). The two SNPs were correlated (r 2=0.79) and regressing out effects of either SNP from the expression levels of ABHD8 and repeating eQTL analysis abolished the eQTL signal for the other SNP, confirming their statistical inseparability. In addition we found significant associations between rs4808616 and NXNL1 expression in OCs (linear regression P=4 × 10−3) and with ANKLE1 expression (P=0.002) in normal ovarian surface epithelial cells (OSECs). There were no eQTL associations for any other genes in the region.
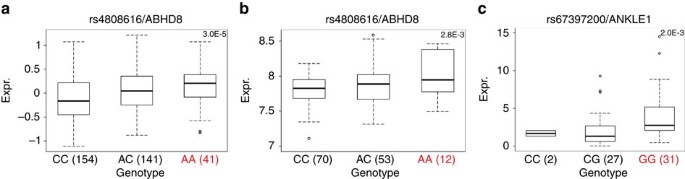
Significant eQTL associations identified between rs4808616 and ABHD8 expression in (a) ovarian cancer tissues and (b) in normal breast tissues. (c) A significant association was also identified between rs4808616 and ANKLE1 expression in primary normal ovarian/fallopian tube epithelial cell cultures. The horizontal line indicates the median expression, the limits of the boxes denote the first and third quartiles, and the whiskers represent 1.5 times the interquartile range of the data. Outliers are indicated with circles.
Full size image
We also performed allele-specific expression analysis in BC using RNA sequencing data31 for coding SNPs in ABHD8 (rs56069439) and BABAM1 (rs10424198). Both SNPs were correlated with rs4808616 (r 2=0.91). There was a significant association between rs56069439 and the allelic ratio of ABHD8 transcripts (F-test P=0.016) with greater expression associated the risk allele (Supplementary Fig. 2; Supplementary Data 3).
Chromosome conformation capture
Chromosome conformation capture (3C) analysis was used to investigate DNA–DNA interactions between ABHD8 and 5 of 13 candidate causal SNPs in Peak 1. Eight SNPs close to the ABHD8 promoter were too near to be resolved, and the close proximity of candidate causal SNPs to ANKLE1 precluded 3C analysis for this gene. The ABHD8 promoter showed an interaction with a 6.3 kb region ∼20 kb telomeric to the gene in both normal breast (Bre80) and ovarian (IOSE11) epithelial cells, and in breast (MCF7) and ovarian (A2780) cancer cell lines (Fig. 3). This region spans the ANKLE1 promoter and includes four candidate causal SNPs: rs4808075, rs10419397, rs56069439 and rs4808076. There was no evidence of interaction for any candidate causal SNP with BABAM1 (Supplementary Fig. 3).
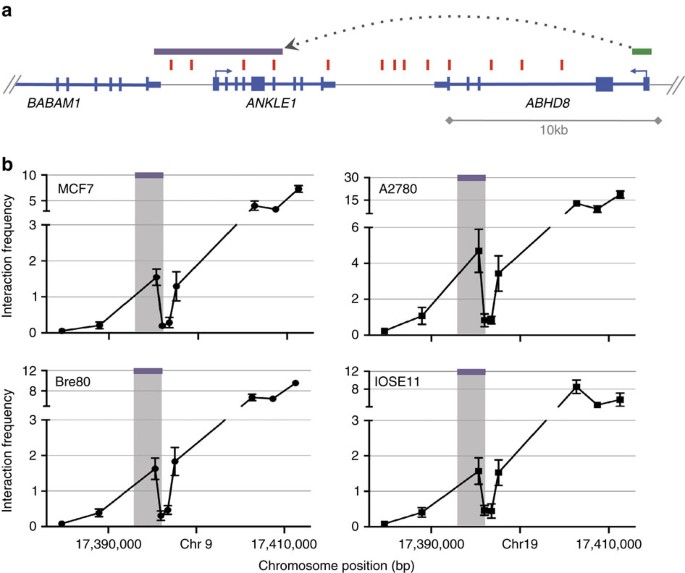
3C interaction profiles in breast and ovarian cell lines. 3C libraries were generated with NcoI, with the anchor point set at the ABHD8 promoter region. (a) A physical map of the region interrogated by 3C is shown, with annotated genes shown in blue, the 13 risk-associated SNPs shown in red, the ABHD8 promoter fragment shown in green and the position of the interacting NcoI fragment represented by the purple bar (not to scale). (b) Relative interaction frequencies between the ABHD8 promoter and regions spanning risk associated SNPs in normal breast (Bre80) and ovarian (IOSE11) epithelial cells lines, and in breast (MCF7) and ovarian (A2780) cancer cell lines. A peak of interaction with the ABHD8 was observed for one region (purple bar) in all four cell lines. There were no interactions detected between the purple region and the BABAM or USHBP1 promoters. The interacting region contains four candidate causal SNPs (from left to right) rs4808075, rs10419397, rs56069439 and rs4808076. Error bars represent s.d. (N=3).
Full size image
Annotation of candidate causal SNPs
All 13 candidate causal SNPs were located in non-protein coding DNA. We annotated putative functional regulatory elements that coincided with the candidate causal SNPs in normal human mammary epithelial cells (HMECs), and normal fallopian tube and ovarian epithelial cells19, and in OC cell lines. Five of the 13 SNPs coincide with regulatory elements that were reproducible in two biological replicate samples (Fig. 4). Three SNPs were located in epigenetic marks in breast and/or ovarian cells: rs55924783 coincided with insulator marks in HMECs and enhancer marks in ovarian cells; rs113299211 coincided with enhancer marks in ovarian cells and is predicted to alter transcription factor binding sites for ELF1, ELK4 and GABP; and rs56069439 coincided with experimentally derived ChIP-seq footprints (for CTCF, ATF2 and ZNF263), enhancer marks in ovarian cells and both enhancer (H3K4me1) and insulator (CTCF) marks in breast cells. Two SNPs were located in 3′-untranslated regions (UTRs) of protein coding genes: rs111961716 in ANKLE1 and rs4808616 in ABHD8. rs4808616 also coincided with enhancer marks in ovarian and breast cells. Finally, rs10419397 lay within the putative promoter of ANKLE1, ∼1,200 bp from the transcription start site.
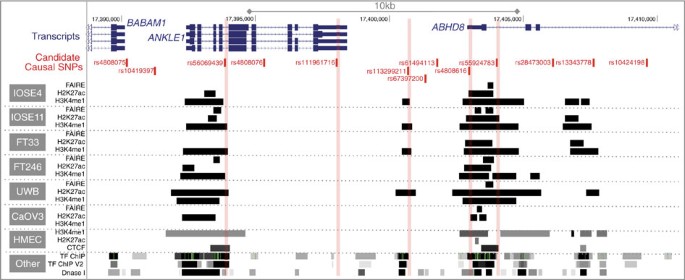
The thirteen candidate SNPs were aligned with open chromatin and enhancer marks (H3K27ac and H3K4me1) in high-grade serous ovarian cancer cells (UWB1.289 and CaOV3) and ovarian cancer precursor cells (ovarian epithelial cells, IOSE and fallopian epithelial cells, FT). Enhancer and insulator (CTCF) data for human mammary epithelial cells (HMECs) were obtained from ENCODE. Five SNPs coincide with biofeatures in breast and/or ovarian cells (indicated in red).
Full size image
Functional analysis of candidate causal SNPs in UTRs
We evaluated the effects on mRNA stability of the SNPs located in 3′ UTRs of ANKLE1 (rs111961716) and ABHD8 (rs4808616, Figs 4 and 5a) in normal primary ovarian epithelial cell lines carrying different SNP genotypes. RNA transcript abundance was measured after blocking mRNA transcription by treating cells with actinomycin D. For rs111961716, ANKLE1 transcript expression was significantly more stable in cell lines homozygous for the A (risk) allele of rs111961716 compared with heterozygous cells or cells homozygous for the C allele (P=0.006, analysis of variance; Fig. 5b). There was no association between ABHD8 mRNA stability and genotypes of rs4808616 (Fig. 5b).
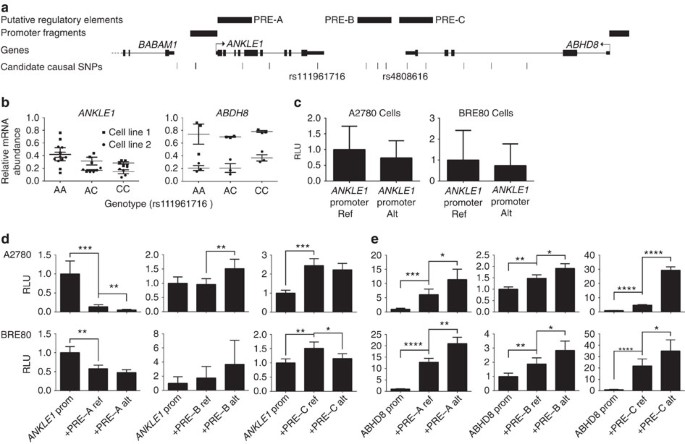
(a) Location of SNPs in putative regulatory elements (PREs) and 5′ untranslated regions. (b) RNA stability assays in primary ovarian epithelial cell lines for risk-associated UTR SNPs in ABHD8 and ANKLE1. Normal ovarian epithelial cell lines carrying different genotypes of the risk SNP rs4808616, located in the 3′ UTR of ABHD8. Rs4808616 is tightly correlated with rs111961716 (R 2=0.98) located in the 3′ UTR of ANKLE1. The risk allele of rs111961716 was associated with decreased mRNA stability of ANKLE1 compared with the protective allele (P=0.006, ANOVA). Different genotypes of rs4808616 are not associated with the stability the ABHD8 transcript. (c–e) Luciferase assays to evaluate SNP-dependent promoter and enhancer activity. (c) The ANKLE1 promoter SNP did not affect ANKLE1 expression in ovarian cancer cells (A2780) and normal breast cells (Bre80). (d) Allele-specific activity of PRE-A, PRE-B and PRE-C on the ANKLE1 promoter. (e) Allele-specific activity of PRE-A, PRE-B and PRE-C on ABHD8 promoter activity. *P>0.05, **P>0.01, ***P>0.001, ****P>0.0001, two-way ANOVA. RLU, relative light units.
Full size image
Functional analysis of promoter and enhancer SNPs
Seven of the 13 candidate causal SNPs in Peak 1 resided either in the ANKLE1 promoter or in putative regulatory elements (PREs-A-C) in breast and ovarian normal and cancer cell lines (Figs 4 and 5a). SNP rs10419397 fell within the ANKLE promoter region, but had no effect on promoter activity (Fig. 5c). PRE-A contained SNP rs56069439, PRE-B contained SNPs rs113299211, rs67397200, rs61494113 and PRE-C contained SNPs rs4808616 and rs55924783. We examined the effect of these PREs, and of the risk alleles of each SNP cloned into luciferase constructs containing the ABHD8 or ANKLE1 promoters. Inclusion of the reference allele of PREs A, B and C significantly increased ABHD8 promoter activity in both OC (A2780) and normal breast (Bre80) cell lines (Fig. 5). Constructs containing the risk alleles further enhanced ABHD8 promoter activity compared with the reference allele for PREs A, B and C in Bre80 cells (P values=0.0027, 0.0308 and 0.0342, respectively, two-way analysis of variance (ANOVA)) and for PREs A, B and C in A2780 cells (P values=0.0193, 0.0115 and <0.0001, respectively, two-way ANOVA; Fig. 5d,e). Constructs containing the reference allele of PRE-A showed a silencing effect on the ANKLE promoter in both cell types with the risk allele further silencing the activity of the reference allele in A2780 cells (P=0.0049, two-way ANOVA). The reference allele of PRE-B had no effect on ANKLE promoter activity, while the risk allele significantly increased activity compared with the reference allele in A2780 cells (P=0.0034, two-way ANOVA). Constructs containing the reference allele of PRE-C significantly increased ANKLE promoter activity in both ovarian (P=0.0004, two-way ANOVA) and breast cell lines (P=0.0067, two-way ANOVA). However the risk allele showed a silencing effect on the reference allele in only Bre80 cells (P=0.0289, two-way ANOVA; Fig. 5d,e).
Functional effects of rs56069439 deletion
Collectively, the data above suggested that rs56069439 may regulate the expression of ANKLE1 and/or ABHD8. We used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated genome editing to delete a 57 bp region containing the regulatory region that includes rs56069439 in breast (MCF10A) and ovarian (IOSE19) epithelial cells (Fig. 6a). Analysis of multiple clones containing confirmed homozygous deletions (Fig. 6b,c) indicated a significant reduction in ANKLE1 expression compared with parental cells (P=0.025, two-tailed paired T-test) and a trend towards reduced ANKLE1 expression in IOSE19 cells (P=0.29, two-tailed paired T-test; Fig. 6d). Expression of ABHD8 and BABAM1 was unchanged following deletion of the region containing rs56069439.
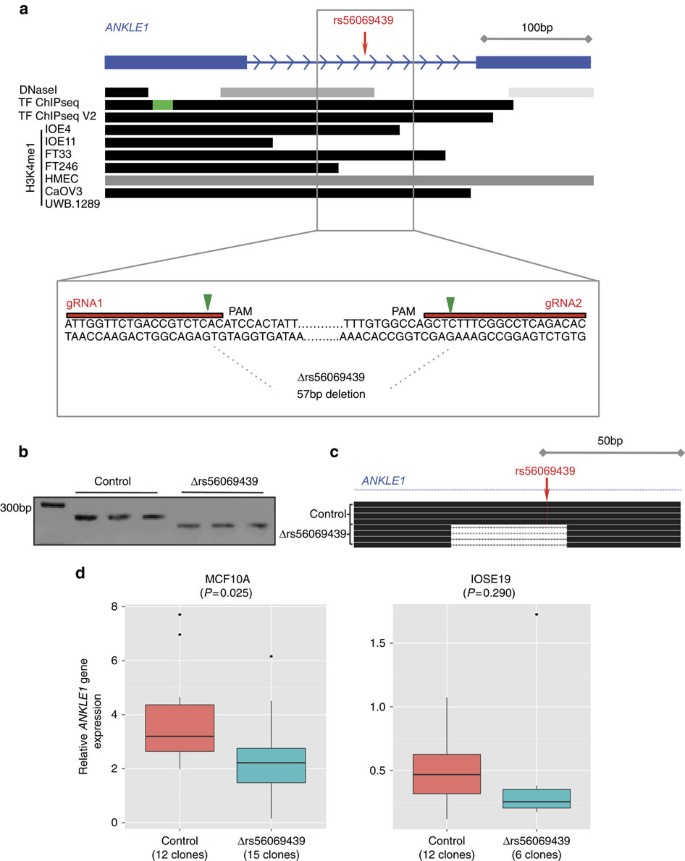
(a) Illustration of the 57 bp region in an intron of ANKLE1 containing rs56069439; H3K4me1 marks overlapped rs56069439 in ovarian, fallopian and breast cells. Location of the two guide RNAs (gRNAs) used to create the stable Δrs56069439 deletion by CRISPR/Cas9 genome editing, cutting sites are indicated with the green arrow. PAM, protospacer adjacent motif. (b) PCR analysis of targeted region in representative MCF10A (breast) epithelial cell clones. Control clones were transfected with the vector backbone only. (c) Verification of deletions by Sanger sequencing, and alignment to the genome using BLAT. (d) Gene expression analysis using TaqMan probes showing downregulation of ANKLE1 was associated with deletion of a region containing rs56069439.
Full size image
In vitro functional analysis of candidate genes
We analysed the effects of perturbing ABHD8, ANKLE1 and BABAM1 expression in in vitro models of 'normal' breast (MCF10A) and ovarian (IOSE19 (ref. 32)) epithelial cells. For each gene, we overexpressed full length, green fluorescent protein-tagged constructs, because genes at 19p13 were frequently overexpressed in ovarian and BCs9 and because eQTL analyses indicated that risk alleles were associated with increased expression of ABHD8 and ANKLE1. After confirming gene overexpression (Supplementary Fig. 3a) we evaluated cell growth, migration and invasion, and anchorage-independent growth (Fig. 7 and Supplementary Fig. 3b). Overexpression of ABHD8 caused a significant reduction in cell migration (P=0.007 in MCF10A; P=0.047 in IOSE19, two-tailed paired T-test) and a decrease in invasion (P=0.018 in MCF10A; P=0.063 in IOSE19, two-tailed paired T-test; Fig. 7). BABAM1 and ANKLE1 overexpression had no effect on these cellular phenotypes for either cell type.
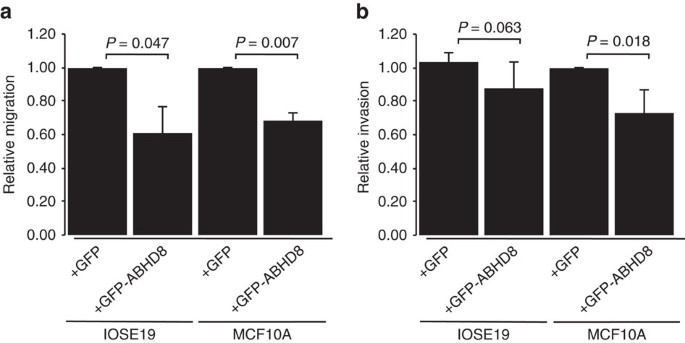
(a) ABHD8 overexpression induced a significant decrease in migration in both breast (MCF10A) and ovarian (IOSE19) cells; (b) ABHD8 overexpression induced a significant decrease in invasion in breast epithelial cells and a similar trend of decreased invasion in ovarian epithelial cells.
Full size image
RNA sequencing was used to profile transcriptomic changes caused by overexpression of ABHD8, ANKLE1 and BABAM1 and pathway analyses performed using Ingenuity Pathway Analysis. We found no indication of significant changes in relevant pathways after overexpressing BABAM1 in breast or ovarian epithelial cells. Cells overexpressing ANKLE1 showed a significant enrichment for cancer-associated and cell growth/proliferation pathways in both breast (P=3.36 × 10−6) and ovarian (P=2.43 × 10−27) epithelial cells. Cells overexpressing ABHD8 were enriched for expression changes in cancer related pathways (P<5.52 × 10−8) and fibrosis pathways (P<1.23 × 10−2, all right-tailed Fisher's exact tests; Supplementary Tables 6-8).
Discussion
Through fine-scale mapping of the 19p13.1 region we have found evidence of two independent regions of genetic association with BC and/or OC risk among women of European ancestry. The minor alleles of all candidate causal variants in Peak 1 conferred increased risks of ER-negative BC and serous OC and increased risks of both cancers for BRCA1 mutation carriers. We were able to rule out associations with ER-positive BC and risks for other OC histotypes. There was weaker evidence that SNPs in Peak 2 were independently associated with BC risk among BRCA1 mutation carriers only. When analyses in BCAC were restricted to triple-negative BC, the strength of association was greater and there was no evidence of association with ER-negative/HER2-positive BC. Thus, our results suggested that these variants are primarily associated with triple-negative BC, the predominant tumour subtype in BRCA1 mutation carriers33. These results are in line with previous findings for the initial SNPs identified through GWAS26.
The increased sample size resulting from combining data from BCAC, OCAC and CIMBA for variants in Peak 1 have enabled us to restrict the likely functional variants at 19p13.1 to 13 SNPs. The 13 candidate causal risk SNPs in this region were the same for both BC and OC leading us to hypothesize that the underlying functional mechanisms are the same in both cancers and the overlap between these SNPs and functional elements provided multiple testable hypotheses, necessitating a range of different functional assays to evaluate their possible causality. Multiple assays were performed in breast and ovarian tissues and cell lines to establish if there is true evidence of pleiotropy. The candidate causal SNPs in Peak 1 clustered around two candidate genes, ANKLE1 and ABHD8, neither of which have been previously implicated in BC or OC. Proximal to these SNPs is BABAM1, a gene involved in recruiting BRCA1 to sites of DNA damage34,35 and therefore a compelling candidate gene at this locus. While gene regulation can be mediated across long genomic distances, the majority of interactions occur over a distance of 1 Mb) or less36,37. We, therefore, evaluated all candidate genes within a 1 Mb region centred on the Peak 1 risk SNPs for eQTL associations. We found significant eQTL associations for ABHD8 in OCs and normal breast tissues, plus allele-specific expression of ABHD8 in BCs, but no compelling evidence for any other gene at this locus. Nonetheless, the identification of ABHD8 as the most likely target susceptibility gene must be treated with some caution as it is plausible that more distant cis-eQTL or even trans-eQTL associations exist for these risk SNPs. Unfortunately, the limited power of eQTL analysis based on the current sample size precluded us from performing genome-wide eQTL analysis to address these hypotheses.
The weight of our functional data, in particular the eQTL associations, indicates that ABHD8 is a target of functional SNPs at this locus, and therefore a novel breast and OC susceptibility gene. 3C identified an interaction between a region containing four candidate causal SNPs and the ABHD8 promoter in both breast and OC and normal epithelial cell lines. The luciferase assays of three PREs (including one encompassing rs56069439 in the interacting region) consistently showed that they acted as enhancers, and furthermore the risk-associated alleles of rs56069439, rs113299211, rs67397200, rs61494113, rs4808616 and rs55924783 (within PREs A-C) further increase ABHD8 promoter activity in both breast and ovarian cells. These results were consistent with our eQTL studies and support the hypothesis that increased ABHD8 expression is associated with an increased cancer risk. ABHD8 is a poorly studied lipase38. The Achilles heel project identified ABHD8 as a lineage-specific cancer cell vulnerability in OC cell lines39 and a recent study identified ABHD8 as a potential OC susceptibility gene though its participation in a homeobox transcription factor-centred gene network associated with serous OC risk40. Overexpression of ABHD8 led to significant reductions in the invasive and migratory potential of breast and ovarian cells and enriched for genes involved in cellular movement (IOSE19) and mTOR signalling (MCF10A), consistent with the observed changes in invasion and migration. The direction of the effect was opposite to what we might expect from the eQTL data, which might reflect different functions of ABHD8 in different contexts, similar to the observations for another BC susceptibility gene, TOX3 (ref. 41). For example, under specific microenvironmental cues or in a tumour cell (rather than the normal cells used in these experiments) increased ABHD8 may promote rather than inhibit migration and invasion.
Nonetheless, we cannot unequivocally exclude other genes as the targets of candidate causal variants at this locus, in particular ANKLE1. The close proximity of the candidate causal SNPs to the ANKLE1 gene precluded 3C analysis; but in the luciferase assays, these same PREs and SNPs had variable, context-dependent effects on ANKLE1 promoter activity. This raises the possibility that the SNPs were cooperatively acting to alter ANKLE1 expression although it was difficult to predict the overall direction of their effects from this assay. We were able to rule out the SNP rs10419397 in the promoter of ANKLE1 as a likely causal variant. The SNP rs111961716 in the 3′-UTR of ANKLE1 was associated with allele-specific ANKLE1 mRNA stability; but stable overexpression of ANKLE1 had no influence on the phenotype of normal breast and ovarian epithelial cells even though pathway after overexpression of ANKLE1 found a significant enrichment for cancer and cell death/proliferation associated pathways in both breast and ovarian epithelial cells. More recently, ANKLE1 has been implicated in DNA damage responses, while other, better-characterized endonucleases (for example, ERCC1) are involved in nucleotide excision repair, which are important for the repair of bulky adducts42.
This study has highlighted the challenges in establishing causality for both candidate causal SNPs at common variant susceptibility loci and the susceptibility genes targets. The multitude of functional assays that can be used to test allele specific functional activity rarely provide unequivocal evidence of one SNP over another. Genome editing, which allows the creation of isogenic experimental models carrying the different alleles of candidate causal SNP, is emerging as a single assay approach that can evaluate the function of common variants. However, until now the technical challenges of genome editing have restricted its application to two non-coding risk SNPs identified by GWAS at susceptibility loci for prostate cancer and obesity, respectively22,23. It was beyond the scope of the current study to utilize genome editing to test all 13 candidate causal SNPs in Peak 1 at 19p13 in BC and OC and normal cell line models. Instead, we used CRISPR-Cas9 genome editing to evaluate the effects of a putative enhancer containing most plausible functional SNP (rs56069439) identified from 3C analysis and mapping of putative regulatory elements. This revealed strong functional evidence for a breast/ovarian epithelial cell enhancer, within an intron of ANKLE1. When this enhancer containing rs56069439 was deleted ANKLE1 expression was significantly reduced, without any reduction in BABAM1 or ABHD8 expression. Further experiments using homology-directed repair will be required to determine if there is allele-specific activity of the rs56069439 SNP in regulating ANKLE1 expression, and to determine whether shadow enhancers are employed to maintain ABHD8 expression43.
In conclusion, we have performed detailed functional analysis of SNPs and candidate target genes at the 19p13 locus in breast and ovarian normal and cancer cells. ABHD8 is the most likely target gene although we cannot rule out a role for ANKLE1 in the development of breast and OC or the possibility that both genes, acting independently or in synergy may be functional targets of candidate causal SNPs. Using a combination of genetic fine mapping, and a spectrum of in silico and functional assays, seven of thirteen showed evidence of functionality.
These data suggest that the underlying functional mechanism(s) at the 19p13 locus may be mediated by many SNPs rather than by a single causal allele. This hypothesis is supported by studies showing tissue-specific enrichment of correlated risk-associated SNPs at susceptibility loci within regulatory biofeatures, including enhancers and transcription factor binding sites19,20. Such enrichments would not be detected if a single causal SNP at a locus was driving disease development. Taken together these data suggest that common molecular mechanisms are likely to underlie this pleiotropic risk locus.
Methods
Study populations
All specimens used in this study were collected with informed consent and under the approval of local Institutional Review Boards. We used epidemiological and genotype data from studies participating in the BCAC44, the OCAC12 and the CIMBA45 that have been genotyped using the iCOGS array that included ∼200,000 SNPs.
BC association consortium
Data were available from 52 BC case-control studies, 41 studies of European ancestry, 9 studies of Asian ancestry and 2 studies of African-American ancestry. Details of all studies, the genotyping process and the quality control process have been described elsewhere6,44, standard sample and genotyping QC criteria were applied. After the quality control process, data on 46,451 cases and 42,599 controls of European ancestry, 6,269 cases and 6,624 controls of Asian ancestry and 1,117 cases and 932 controls of African-American ancestry were available for analysis. Data on the BC ER status were available for 34,509 cases of European ancestry, 7,435 (22%) of whom had ER-negative tumours.
OC association consortium
Data were available from 41 case-control studies of EOC from OCAC that were genotyped using the iCOGS array12. In addition to the OCAC iCOGS data, genotype data were available for stage 1 of three population-based OC genome-wide association studies. The final data set comprised genotype data for 11,069 cases and 21,722 controls from COGS ('OCAC-iCOGS'), 2,165 cases and 2,564 controls from a GWAS from North America ('US GWAS')46, 1,762 cases and 6,118 controls from a UK-based GWAS ('UK GWAS')7, and 441 cases and 441 controls from the Mayo Clinic. All subjects included in this analysis provided written informed consent as well as data and blood samples under ethically approved protocols. Overall, 43 studies from 11 countries provided data on 15,437 women diagnosed with invasive EOC, 9,627 of whom were diagnosed with serous EOC and 30,845 controls from the general population.
Consortium of investigators of modifiers of BRCA1/2
Data on BRCA1 mutation carriers were obtained through CIMBA. Eligibility in CIMBA is restricted to females 18 years or older with pathogenic mutations in BRCA1 or BRCA2. The majority of the participants were sampled through cancer genetics clinics47, including some related participants. Fifty-one studies from 25 countries contributed data on BRCA1 mutation carriers who were genotyped using the iCOGS array45. After quality control of the phenotypes and genotypes, data were available on 15,252 BRCA1 mutation carriers of whom 7,455 had been diagnosed with BC, 2,639 with ER-negative BC and 1,724 with OC, all of European ancestry. Analyses in BRCA1 mutation carriers focused on assessing associations with BC risk, following the evidence from the original GWAS in BRCA1 mutation carriers48.
URLs: 1000 Genomes Project, http://www.1000genomes.org/; BCAC, http://ccge.medschl.cam.ac.uk/consortia/bcac/index.html; CIMBA, http://ccge.medschl.cam.ac.uk/consortia/cimba/index.html; COGS, http://www.cogseu.org/; iCOGS, http://ccge.medschl.cam.ac.uk/research/consortia/ icogs/; SNAP
https://www.broadinstitute.org/mpg/snap/; TCGA, https://tcga-data.nci.nih.gov; CGHub, https://cghub.ucsc.edu/
iCOGS SNP selection for fine mapping and imputation
The fine mapping region was defined as Chromosome 19 positions: 17,130,000–17,550,000 (NCBI build 37). To identify the set of variants potentially responsible for the original GWAS reports, we considered all variants with minor allele frequencies of >0.02 from the 1,000 Genomes Project (March 2010 version) and selected all SNPs correlated (r 2>0.1) with either of the two SNPs that had been identified through the BRCA1 and EOC GWAS studies (rs8170 and rs2363956)12,45, plus an additional set of SNPs that tagged all remaining SNPs in the region with r 2>0.9. A total of 438 SNPs that were included on iCOGS in the 19p13 region passed QC and were available for the analyses. Data on these SNPs were used to impute the genotypes of all known variants from the 1,000 genomes project (V3, April 2012 release49) using the IMPUTE (version 2) software. After excluding SNPs with MAF<0.001 and SNPs with imputation r 2 accuracy score of ≤0.3, there were 2,269 imputed SNPs in BCAC, 2,565 in OCAC and 2,311 in BRCA1 mutation.
BCAC and OCAC association analysis and logistic regression
To evaluate the association of each SNP with breast and EOC risk in BCAC and OCAC we used a Wald test statistic based on logistic regression, by estimating the per-allele OR and its s.e. Analyses restricted to specific tumour subtypes (ER-negative BC or high-grade serous EOC) were assessed separately using all available controls. All analyses were adjusted for principal components, described in more detail elsewhere12,44. Conditional logistic regression was used to assess the evidence that there are multiple independent association signals in the region, by evaluating the associations of genetic variants in the region while adjusting for the SNP with the smallest P value. We considered only SNPs with P values of association of <10−3 and MAF>0.1% and the most parsimonious model was identified using step-wise forward logistic regression and a threshold of P<10−4 for retaining SNPs in the model.
CIMBA retrospective cohort analysis
All associations between genotypes and BC risk in BRCA1 mutation carriers were evaluated using a 1 df per allele trend-test (P-trend), based on modelling the retrospective likelihood of the observed genotypes conditional on BC phenotypes49. To allow for the non-independence among related individuals, an adjusted test statistic was used which took into account the correlation in genotypes48. Per allele HR estimates were obtained by maximizing the retrospective likelihood. All analyses were stratified by country of residence. To identify the most parsimonious model that includes multiple SNPs, forward-selection Cox-regression analysis was performed, using the same P value thresholds as in the BCAC and OCAC analysis. This approach provides valid tests of association, although the parameter estimates can be biased49,50. Parameter estimates for the most parsimonious model were obtained using the retrospective likelihood approach.
Meta-analysis
It is well established that the majority of BCs in BRCA1 mutation carriers are ER-negative51,52. To increase the statistical power for identifying the most likely causal variants, we also performed a meta-analysis of the associations of BC risk for BRCA1 mutation carriers and ER-negative BC in the general population (in BCAC) for both genotyped and imputed SNPs. We used an inverse variance approach assuming mixed effects, by combining the logarithm of the per-allele HR for the association with BC risk for BRCA1 mutation carriers and the logarithm of the OR estimate for the association with ER-negative BC in BCAC.
eQTL and allele-specific expression analyses
Germline genotype data were obtained from the Affymetrix SNP 6.0 (METABRIC) and Illumina 1M-Duo (TCGA HGSOC). No SNPs from Peak 1 and 2 were present on the Affymetrix platform so these genotypes were imputed into the 1000 Genomes European reference panel (March 2012, version 3) using IMPUTE version 2 (ref. 53). All analyses were restricted to patients of >90% European ancestry as per LAMP estimates54 and SNPs with info score >0.3. For METABRIC, gene expression data consisted of probe-level measurements from the Illumina HT-12 v3 microarray platform for a total of 135 samples obtained from normal breast tissue adjacent to tumour and 59 samples obtained from ER-negative breast tumours were analysed. For TCGA HGSOC, gene expression data consisted of measurements from the Agilent 244 K microarray for 340 HGSOC tumours downloaded from the cBioportal. Only genes and probes <1 Mb from the top Peak 1 SNP were analysed. Tumour gene expression data was first adjusted for copy number (TCGA and METBRIC, Affymetrix SNP 6.0 calls) and methylation (TCGA only, Illumina 27 K beta values) using the method of Li et al 31. Expression QTL analysis was conducted by linear regression with genotypes as predictors, as implemented in the R package Matrix eQTL55.
Sixty early passage primary normal OSECs and fallopian tube epithelial cells were collected and cultured as previously described27,56. Briefly, OSECs were harvested from ovaries using a sterile cytobrush and cultured in Medium 199 and MCDB105, mixed in a 1:1 ratio and supplemented with 15% fetal bovine serum (FBS, Hyclone), 10 ng ml−1 epidermal growth factor, 0.5 mg ml−1 hydrocortisone, 5 mg ml−1 insulin (all Sigma, St Louis, MO, USA) and 34 mg protein per ml bovine pituitary extract (Life Technologies). Fresh fallopian specimens were subjected to 48–72 h Pronase (Roche) and DNase I digests to release the epithelial cells. Epithelial cells were pelleted and cultured on collagen in DMEM/F12 supplemented with 10% FBS (Seradigm). RNA was isolated from cell cultures harvested at ∼80% confluency using the QIAgen miRNAeasy kit with on-column DNase 1 digestion. 500 ng of RNA was reverse transcribed using SuperScript III First-Strand Synthesis System (Invitrogen). The cDNA was diluted to 10 ng μl−1 and 12.5 ng was used in target specific amplification before real-time PCR using TaqMan PreAmp Master Mix Kit (Applied Biosystems) following Fluidigm's Specific Target Amplification Protocol. 1.25 μl of the 25 μl pre-amplified cDNA was added to each chip. Each sample was run in triplicate and each experiment included no template controls and no template controls from the cDNA reactions. 96.96 Dynamic Array Integrated Fluidic Circuits (Fluidigm) were loaded with 96 pre-amplified cDNA samples and 96 TaqMan gene expression probes (Applied Biosystems) using the BioMark HD System (Fluidigm). Expression levels for each gene were normalized to the average expression of control genes (GAPDH and ACTB). Relative expression levels were calculated using the ΔΔCt method. Correlations between genotype and gene expression were calculated in R 2.14.1. Genotype specific gene expression was compared using the Jonckheere–Terpstra test. Genes with significant eQTL results were validated by individual Taqman (Applied Biosystems, Warrington UK) reactions run on ABI 7900HT Sequence Detection System equipment and analysed with SDS software according to the manufacturer's instructions. Normal cell line DNAs were analysed on iCOGS arrays to obtain genotype information. We analysed all protein-coding genes within a 1 Mb region of the risk association. The method for allele specific expression analysis has been described previously31.
Breast and ovarian normal and cancer cell lines
Breast and OC cell lines MCF7 (ER+, breast; ATCC #HTB-22) and A2780 (ER+, ovarian; kindly provided by Thomas Hamilton, NCI, Maryland) were grown in RPMI medium with 10% FBS and antibiotics. The normal breast epithelial cell lines Bre-80 (kindly provided by Roger Reddel, CMRI, Sydney) and MCF10A (ATCC #CRL-10317) were grown in DMEM/F12 medium with 5% horse serum, 10 mg ml−1 insulin, 0.5 mg ml−1 hydrocortisone, 20 ng ml−1 epidermal growth factor, 100 ng ml−1 cholera toxin and antibiotics. The phenotypically normal TERT immortalized ovarian epithelial cell lines IOSE11 and IOSE19 (ref. 32) were grown in NOSE-CM. All cell lines were maintained under standard conditions, were routinely tested for Mycoplasma and were profiled with short tandem repeats to confirm their identity.
Functional annotation of risk SNPs
FAIRE-seq and ChIP-seq for H3K27ac and H3K4me1 marks in normal ovarian (IOSE4, IOSE11) and fallopian epithelial cell lines (FT33, FT246) and OC cell lines (CaOV3, UWB1.289) were generated in-house using standard protocols and have been previously described19,27. Epigenetic marks in HMECs were downloaded from ENCODE (genome.ucsc.edu).
Chromosome conformation capture
3C libraries were generated using NcoI as described previously14. To quantify interactions by real-time quantitative PCR (qPCR) was performed using primers listed in Supplementary Table 9. All qPCRs were performed on a RotorGene 6,000 using MyTaq HS DNA polymerase with the addition of 5 mM of Syto9, annealing temperature of 66 °C and extension of 30 s. Each experiments was performed three times in duplicate. The BAC clone (CTD-2278I10) covering the 19p13 region was used to normalize for PCR efficiency and a by reference region within GAPDH used to calculate relative interaction frequencies. All qPCR products were resolved on 2% agarose gels, gel purified and sequenced to verify the 3C product.
RNA stability assays
For each genotype (two homozygotes and the heterozygote) two early passage primary normal ovarian epithelial cell lines were incubated with actinomycin D for 20 h. RNA was extracted using the QIAgen RNeasy extraction kit and reverse transcribed using MMLV RT enzyme and random hexamers (Promega). Quantitative PCR was performed using TaqMan gene expression probes for ABHD8 (Hs00225984_m1) and ANKLE1 (Hs01094673_g1). Signal for each gene of interest was normalized to signal for ACTB (Hs01060665_g1) and GAPDH (Hs02758991_g1) and relative gene expression calculated using the ΔΔCt method, relative to untreated cells. 18s rRNA (Hs99999901_s1) and MYC (Hs00153408_m1) mRNA levels were included as internal controls.
Promoter and allele specific enhancer assays
A 1119, bp fragment containing the ABHD8 promoter was cloned into the pGL3 basic luciferase reporter. Reference and risk associated ANKLE1 promoter fragments were synthesized by GenScript and cloned into pGL3 basic. We generated PCR fragments corresponding to PRE A and PRE B and had PRE C haplotype fragments synthesized by GenScript and these were also sub-cloned into ABHD8 and ANKLE1 promoter constructs. PCR primers are listed in Supplementary Table 10. Bre80 and A2780 cells were transiently transfected with equimolar amounts of luciferase reporter constructs using Renilla luciferase as an internal control reporter. Luciferase was measured 24 h after transfection using Dual-Glo Luciferase (Promega). To correct for any differences in transfection efficiency or cell lysate preparation, Firefly luciferase activity was normalized to Renilla luciferase, and the activity of each construct was measured relative to the promoter alone construct, which had a defined activity of 1. Association was assessed by log transforming the data and performing two-way ANOVA, followed by Dunnett's multiple comparisons test; for ease of interpretation, values were back transformed to the original scale for the graphs.
Genome editing
Guide RNAs targeting the region flanking rs56069439 (5′-GTGAGACGGTCAGAACCAAT-3′ and 5′-GTGTCTGAGGCCGAAAGAGC-3′) were designed using the CRISPR design tool from the Zhang lab (www.crispr.mit.edu)57. The gRNAs were cloned into the lentiCRISPR (Addgene Plasmid 49535) vector by using the BsmBI restriction enzyme site and lentiviral supernatants made by cotransfection of HEK293T cells. IOSE19 and MCF10A cells were transduced with viral supernatants and infected cells selected using 400 ng ml−1 and 500 ng ml−1 puromycin (Sigma Aldrich) respectively. Selected cells were sorted into single cells using flow cytometry and expanded in vitro. Screening for clones containing the deletion was performed using the following primers: Forwards: 5′-CCCTGACATCCAGGGTCTTC-3′ and Reverse: 5′-AGTCCAGCGTCTCATCGGTA-3′. For sequence verification of the deletion the following primers were used: Forwards: 5′-TTCTGGACCAGTCCCTGACA-3′ and Reverse: 5′-CAGCGTCTCATCGGTAGGTC-3′. RNA was isolated from positive clones using the Zymo Quick-RNA kit and reverse transcribed using Superscript III (Life Technologies). Real time gene expression analysis was performed using TaqMan probes, as described above.
In vitro analysis of candidate genes
The three candidate genes were overexpressed as green fluorescent protein fusion proteins. The BABAM1 overexpression construct was a kind gift from Dr S Elledge58. ANKLE1 and ABHD8 contructs were purchased from Genecopoeia. Virus was made in-house by cotransfection of HEK293Ts and used to transduce MCF10A and IOSE19 cells. Positive cells were selected using 400 ng ml−1 (for IOSE19 cells) or 500 ng ml−1 (for MCF10A cells) puromycin. Anchorage dependent and independent growth assays were performed as previously described32,59. For invasion and migration assays Millipore luminescent transwell assays (24 well plate format) were used, following the manufacturer's protocol.
Data availability
The relevant SNP genotype data underpinning these analyses can be accessed by applying to the OCAC, BCAC and CIMBA consortia (see URLs). EQTL data are available in supplementary information. All other data are available on request.
Additional information
How to cite this article: Lawrenson, K. et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun. 7:12675 doi: 10.1038/ncomms12675 (2016).
References
- 1
Easton, D. F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 (2007).
CAS ADS Article Google Scholar
- 2
Ahmed, S. et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 41, 585–590 (2009).
CAS Article Google Scholar
- 3
Ghoussaini, M. et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 44, 312–318 (2012).
CAS Article Google Scholar
- 4
Siddiq, A. et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21, 5373–5384 (2012).
CAS Article Google Scholar
- 5
Garcia-Closas, M. et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 45, 392–398 398e1-2 (2013).
CAS Article Google Scholar
- 6
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361 361e1-2 (2013).
CAS Article Google Scholar
- 7
Song, H. et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 41, 996–1000 (2009).
CAS Article Google Scholar
- 8
Goode, E. L. et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 42, 874–879 (2010).
CAS Article Google Scholar
- 9
Bolton, K. L. et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 42, 880–884 (2010).
CAS Article Google Scholar
- 10
Bojesen, S. E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 45, 371–384 384e1-2 (2013).
CAS Article Google Scholar
- 11
Permuth-Wey, J. et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat. Commun. 4, 1627 (2013).
Article Google Scholar
- 12
Pharoah, P. D. et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45, 362–370 370e1-2 (2013).
CAS Article Google Scholar
- 13
Shen, H. et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat. Commun. 4, 1628 (2013).
Article Google Scholar
- 14
Ghoussaini, M. et al. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 4, 4999 (2014).
Article Google Scholar
- 15
Meyer, K. B. et al. Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am. J. Hum. Genet. 93, 1046–1060 (2013).
CAS Article Google Scholar
- 16
French, J. D. et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 92, 489–503 (2013).
CAS Article Google Scholar
- 17
Orr, N. et al. Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2. Hum. Mol. Genet. 24, 2966–2984 (2015).
CAS Article Google Scholar
- 18
Glubb, D. M. et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am. J. Hum. Genet. 96, 5–20 (2015).
CAS Article Google Scholar
- 19
Coetzee, S. G. et al. Cell type specific enrichment of risk associated regulatory elements at ovarian cancer susceptibility loci. Hum. Mol. Genet. 24, 3595–3607 (2015).
CAS Article Google Scholar
- 20
Hazelett, D. J. et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 10, e1004102 (2014).
Article Google Scholar
- 21
Cowper-Sal lari, R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44, 1191–1198 (2012).
CAS Article Google Scholar
- 22
Spisák, S. et al. CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat. Med. 21, 1357–1363 (2015).
Article Google Scholar
- 23
Claussnitzer, M. et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 373, 895–907 (2015).
CAS Article Google Scholar
- 24
Antoniou, A. C. et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 42, 885–892 (2010).
CAS Article Google Scholar
- 25
Couch, F. J. et al. Common variants at the 19p13.1 and ZNF365 loci are associated with ER subtypes of breast cancer and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol. Biomarkers Prev. 21, 645–657 (2012).
CAS Article Google Scholar
- 26
Stevens, K. N. et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 72, 1795–1803 (2012).
CAS Article Google Scholar
- 27
Kuchenbaecker, K. B. et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 47, 164–171 (2015).
CAS Article Google Scholar
- 28
Stevens, K. N. et al. Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res. 71, 6240–6249 (2011).
CAS Article Google Scholar
- 29
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
CAS Article Google Scholar
- 30
Network, C. G. A. R. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
- 31
Li, Q. et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152, 633–641 (2013).
CAS Article Google Scholar
- 32
Lawrenson, K. et al. Senescent fibroblasts promote neoplastic transformation of partially transformed ovarian epithelial cells in a three-dimensional model of early stage ovarian cancer. Neoplasia 12, 317–325 (2010).
CAS Article Google Scholar
- 33
Mavaddat, N. et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol. Biomarkers Prev. 21, 134–147 (2012).
CAS Article Google Scholar
- 34
Shao, G. et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
CAS Article Google Scholar
- 35
Hu, X. et al. NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes. J. Biol. Chem. 286, 11734–11745 (2011).
CAS Article Google Scholar
- 36
Stranger, B. E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012).
CAS Article Google Scholar
- 37
Sanyal, A., Lajoie, B.R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
CAS ADS Article Google Scholar
- 38
Thomas, G., Brown, A. L. & Brown, J. M. In vivo metabolite profiling as a means to identify uncharacterized lipase function: recent success stories within the alpha beta hydrolase domain (ABHD) enzyme family. Biochim. Biophys. Acta 1841, 1097–1101 (2014).
CAS Article Google Scholar
- 39
Cheung, H. W. et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl Acad. Sci. USA 108, 12372–12377 (2011).
CAS ADS Article Google Scholar
- 40
Kar, S. P. et al. Network-Based Integration of GWAS and Gene Expression Identifies a HOX-Centric Network Associated with Serous Ovarian Cancer Risk. Cancer Epidemiol. Biomarkers Prev. 24, 1574–1584 (2015).
CAS Article Google Scholar
- 41
Seksenyan, A. et al. TOX3 is expressed in mammary ER(+) epithelial cells and regulates ER target genes in luminal breast cancer. BMC Cancer 15, 22 (2015).
Article Google Scholar
- 42
Selvakumaran, M., Pisarcik, D. A., Bao, R., Yeung, A. T. & Hamilton, T. C. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 63, 1311–1316 (2003).
CAS PubMed PubMed Central Google Scholar
- 43
Frankel, N. et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010).
CAS ADS Article Google Scholar
- 44
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361 361e1–2 (2013).
CAS Article Google Scholar
- 45
Couch, F.J. et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 9, e1003212 (2013).
CAS Article Google Scholar
- 46
Permuth-Wey, J. et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 71, 3896–3903 (2011).
CAS Article Google Scholar
- 47
Chenevix-Trench, G. et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res. 9, 104 (2007).
Article Google Scholar
- 48
Antoniou, A. C. et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 42, 885–892 (2010).
CAS Article Google Scholar
- 49
Barnes, D. R., Lee, A., Easton, D. F. & Antoniou, A. C. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet. Epidemiol. 36, 274–291 (2012).
Article Google Scholar
- 50
Antoniou, A. C. et al. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet. Epidemiol. 29, 1–11 (2005).
Article Google Scholar
- 51
Mavaddat, N. et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol. Biomarkers Prev. 21, 134–147 (2012).
CAS Article Google Scholar
- 52
Lakhani, S. R. et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin.Oncol. 20, 2310–2318 (2002).
CAS Article Google Scholar
- 53
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Article Google Scholar
- 54
Baran, Y. et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28, 1359–1367 (2012).
CAS Article Google Scholar
- 55
Shabalin, A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012).
CAS Article Google Scholar
- 56
Karst, A. M. & Drapkin, R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nat. Protoc. 7, 1755–1764 (2012).
CAS Article Google Scholar
- 57
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
CAS Article Google Scholar
- 58
Wang, B., Hurov, K., Hofmann, K. & Elledge, S. J. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 23, 729–739 (2009).
CAS Article Google Scholar
- 59
Lawrenson, K. et al. Modelling genetic and clinical heterogeneity in epithelial ovarian cancers. Carcinogenesis 32, 1540–1549 (2011).
CAS Article Google Scholar
Download references
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out, in particular those involved in the COGS project: Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, the staff of the CNIO genotyping unit, Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility.
BCAC (acknowledgements by study) (ABCFS): Maggie Angelakos, Judi Maskiell, Gillian Dite. (ABCS) C Ellen van der Schoot, Sanquin Amsterdam. (ACP) The ACP study wishes to thank the participants in the Thai Breast Cancer study. Special Thanks also go to the Thai Ministry of Public Health (MOPH), doctors and nurses who helped with the data collection process. Finally, the study would like to thank Dr Prat Boonyawongviroj, the former Permanent Secretary of MOPH and Dr Pornthep Siriwanarungsan, the Department Director-Generalof Disease Control who have supported the study throughout. (BBCS) Eileen Williams, Elaine Ryder-Mills, Kara Sargus (BIGGS) Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones. (BSUCH) Peter Bugert, Medical Faculty Mannheim (CGPS) Staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen (CNIO-BCS) Guillermo Pita, Charo Alonso, Daniel Herrero, Nuria Álvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO)(CTS). The CTS Steering Committee includes Leslie Bernstein, Susan Neuhausen, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu, and Jessica Clague DeHart at the Beckman Research Institute of City of Hope, Dennis Deapen, Rich Pinder, Eunjung Lee, and Fred Schumacher at the University of Southern California, Pam Horn-Ross, Peggy Reynolds, Christina Clarke Dur and David Nelson at the Cancer Prevention Institute of California, and Hoda Anton-Culver, Argyrios Ziogas, and Hannah Park at the University of California Irvine. (ESTHER) Hartwig Ziegler, Sonja Wolf, Volker Hermann. (GC-HBOC) Heide Hellebrand, Stefanie Engert and GC-HBOC (Supported by Deutsche Krebshilfe). (GENICA) The GENICA Network: Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [HB, Wing-Yee Lo, Christina Justenhoven], German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ) [HB], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [UH], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth](HEBCS) Kirsimari Aaltonen, Karl von Smitten, Sofia Khan, Tuomas Heikkinen, Irja Erkkilä. (HMBCS) Natalia Antonenkova, Peter Hillemanns, Hans Christiansen and Johann H. Karstens (KBCP) Eija Myöhänen, Helena Kemiläinen. (kConFab/AOCS) We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. (LAABC) We thank all the study participants and the entire data collection team, especially Annie Fung and June Yashiki. (LMBC) Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts. (MARIE) Petra Seibold, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. (MCCS) MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index. (MBCSG) Bernard Peissel and Daniela Zaffaroni and Giulietta Scuvera of the Fondazione IRCCS Istituto Nazionale dei Tumori (INT), Milan, Italy; Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia (IEO), Milan, Italy; and the personnel of the Cogentech Cancer Genetic Test Laboratory. (MTLGEBCS) We would like to thank Martine Tranchant (CHU de Québec Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management and skilful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. (MYBRCA) Phuah Sze Yee, Peter Kang, Kang In Nee, Kavitta Sivanandan, Shivaani Mariapun, Yoon Sook-Yee, Daphne Lee, Teh Yew Ching and Nur Aishah Mohd Taib for DNA Extraction and patient recruitment. (NBHS) We thank study partcipants and research staff for their contributions and commitment to this study. (OBCS) Meeri Otsukka, Kari Mononen(OFBCR) Teresa Selander, Nayana Weerasooriya(ORIGO) We thank E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr J. Molenaar. (PBCS) Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner(pKARMA) The Swedish Medical Research Counsel. (RBCS) Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk, the Erasmus MC Family Cancer Clinic. (SASBAC) The Swedish Medical Research Counsel. (SBCGS) We thank study partcipants and research staff for their contributions and commitment to this study. (SBCS) Sue Higham, Helen Cramp, Ian Brock, Malcolm W. R. Reed, Sabapathy Balasubramanian and Dan Connley. (SEARCH) The SEARCH and EPIC teams. (SGBCC) We thank the participants and research coordinator Kimberley Chua. (SKKDKFZS) We thank all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. (TNBCC) Robert Pilarski and Charles Shapiro were instrumental in the formation of the OSU Breast Cancer Tissue Bank. We thank the Human Genetics Sample Bank for processing of samples and providing OSU Columbus area control samples. (UKBGS) We thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre.
OCAC (acknowledgements by study): This study would not have been possible without the contributions of the following: J Dennis, P. Hall (COGS); D. C. Tessier, F. Bacot, D. Vincent, S. LaBoissière and F. Robidoux and the staff of the genotyping unit, (Genome Quebec); D. C. Whiteman, P. M. Webb, A. C. Green, N. K. Hayward, P. G. Parsons, D. M. Purdie, B. M. Smithers, D. Gotley, A. Clouston, I. Brown, S. Moore. K. Harrap, T. Sadkowski, S. O'Brien, E. Minehan, D. Roffe, S. O'Keefe, S. Lipshut, G. Connor, H. Berry, F. Walker, T. Barnes, J. Thomas, L. Terry, M. Connard, L. Bowes, M-R. Malt, J. White, C. Mosse, N. Tait, C. Bambach, A. Biankan, R. Brancatisano, M. Coleman, M. Cox, S. Deane, G. L. Falk, J. Gallagher, M. Hollands, T. Hugh, D. Hunt, J. Jorgensen, C. Martin, M. Richardson, G. Smith, R. Smith, D. Storey, J. Avramovic, J. Croese, J. D'Arcy, S. Fairley, J. Hansen, J. Masson, L. Nathanson, B. O'Loughlin, L. Rutherford, R. Turner, M. Windsor, J. Bessell, P. Devitt, G. Jamieson, D. Watson, S. Blamey, A. Boussioutas, R. Cade, G. Crosthwaite, I. Faragher, J. Gribbin, G. Hebbard, G. Kiroff, B. Mann, R. Millar, P. O'Brien, R. Thomas, S. Wood, S. Archer, K. Faulkner, J. Hamdorf (ACS); R. Stuart-Harris, F. Kirsten, J. Rutovitz, P. Clingan, A.Glasgow, A. Proietto, S. Braye, G. Otton, J. Shannon, T. Bonaventura, J. Stewart, S. Begbie, M. Friedlander, D. Bell, S. Baron-Hay, G. Gard, D. Nevell, N. Pavlakis, S. Valmadre, B. Young, C Camaris, R. Crouch, L. Edwards, N. Hacker, D. Marsden, G. Robertson, P. Beale, J. Beith, J. Carter, C. Dalrymple, R. Houghton, P. Russell, L. Anderson, M. Links, J. Grygiel, J. Hill, A. Brand, K. Byth, R. Jaworski, P. Harnett, R. Sharma,.G Wain, D. Purdie, D. Whiteman, B. Ward, D. Papadimos, A. Crandon, M. Cummings, K. Horwood. A. Obermair, L. Perrin, D. Wyld, J. Nicklin, M. Davy, M. K. Oehler, C. Hall, T. Dodd, T. Healy, K. Pittman, D. Henderson, J. Miller, J. Pierdes, A. Achan, P. Blomfield, D. Challis, R. McIntosh, A. Parker, B. Brown, R. Rome, D. Allen, P. Grant, S. Hyde, R. Laurie, M. Robbie, D. Healy, T. Jobling, T. Manolitsas, J. McNealage, P Rogers, B. Susil, E. Sumithran, I. Simpson, I. Haviv, K. Phillips, D. Rischin, S. Fox, D. Johnson, S. Lade, P. Waring, M. Loughrey, N.O'Callaghan, B. Murray, L. Mileshkin, P. Allan; V. Billson, J. Pyman, D. Neesham, M. Quinn, A. Hamilton, C. Underhill, R. Bell, L. F Ng, R. Blum, V.Ganju, I. Hammond, C. Stewart, Y. Leung, M. Buck, N. Zeps (ACS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); U. Eilber and T. Koehler (GER); L. Gacucova (HMO); P. Schürmann, F. Kramer, W. Zheng, T.-W. Park-Simon, K. Beer-Grondke and D. Schmidt (HJO); G.S. Keeney, S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL (NHS); L. Paddock, M. King, U. Chandran, A. Samoila, and Y. Bensman (NJO); L. Brinton, M. Sherman, A. Hutchinson, N. Szeszenia- Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); ); C. Luccarini, P. Harrington the SEARCH team and ECRIC (SEA); the Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators (SRO); W-H. Chow, Y-T. Gao (SWH); Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/ (TCGA); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); Carole Pye (UKR); a full list of the investigators who contributed to the generation of the WTCCC data is available from http://www.wtccc.org.uk/ (WTCCC).
CIMBA (acknowledgements by study): (BCFR-AU) Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis. (BCFR-NY) We wish to thank members and participants in the New York site of the Breast Cancer Family Registry for their contributions to the study. (BCFR-ON) We wish to thank members and participants in the Ontario Familial Breast Cancer Registry for their contributions to the study. (BFBOCC-LT) We acknowledge Vilius Rudaitis, Laimonas Griškevičius, Ramūnas Janavičius (if not in the authorship). BFBOCC-LV acknowledge Drs Janis Eglitis, Anna Krilova and Aivars Stengrevics. (BMBSA) We wish to thank the families who contribute to the BMBSA study. (BRICOH) We wish to thank Yuan Chun Ding and Linda Steele for their work in participant enrolment and biospecimen and data management.(CNIO) We thank Alicia Barroso, Rosario Alonso and Guillermo Pita for their assistance. (CONSIT TEAM) Alessandra Viel and Riccardo Dolcetti of the CRO Aviano National Cancer Institute, Aviano (PN), Italy; Laura Ottini of the 'Sapienza' University, Rome, Italy; Liliana Varesco of the IRCCS AOU San Martino - IST Istituto Nazionale per la Ricerca sul Cancro, Genoa, Italy; Laura Papi and Gabriele Capone of the University of Florence, Florence, Italy; Antonella Savarese and Aline Martayan of the Istituto Nazionale Tumori Regina Elena, Rome, Italy; Stefania Tommasi and Brunella Pilato of the Istituto Nazionale Tumori 'Giovanni Paolo II', Bari, Italy. (CORE) The CIMBA data management and analysis is funded through Cancer Research- UK grant C12292/A11174. ACA is a Senior Cancer Research - UK Research Fellow. (EMBRACE) RE is supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. (FCCC) We thank Ms. JoEllen Weaver and Dr Betsy Bove for their technical support. (GEMO) Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) study: National Cancer Genetics Network «UNICANCER Genetic Group», France. We wish to thank all the GEMO collaborating groups for their contribution to this study. GEMO Collaborating Centres are: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon - Centre Léon Bérard, and Equipe «Génétique du cancer du sein», Centre de Recherche en Cancérologie de Lyon: Olga Sinilnikova†, Sylvie Mazoyer, Francesca Damiola, Laure Barjhoux, Carole Verny-Pierre, Mélanie Léone, Nadia Boutry-Kryza, Alain Calender, Sophie Giraud; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Etienne Rouleau, Lisa Golmard, Agnès Collet, Virginie Moncoutier, Cédrick Lefol, Muriel Belotti, Antoine de Pauw, Camille Elan, Catherine Nogues, Emmanuelle Fourme, Anne-Marie Birot. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Olivier Caron, Marine Guillaud-Bataille. Centre Jean Perrin, Clermont–Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona, Sandrine Handallou. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet, Dominique Vaur, Laurent Castera. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, Audrey Remenieras, François Eisinger. CHU Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin†, Claude Adenis. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon. CHU Dijon: Fanny Coron, Laurence Faivre. CHU St-Etienne: Fabienne Prieur, Marine Lebrun, Caroline Kientz. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU Limoges: Laurence Vénat-Bouvet. CHU Nantes: Capucine Delnatte. CHU Bretonneau, Tours: Isabelle Mortemousque. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier, Mathilde Warcoin. CHU Vandoeuvre-les-Nancy: Johanna Sokolowska, Myriam Bronner. CHU Besançon: Marie-Agnès Collonge-Rame, Alexandre Damette. Creighton University, Omaha, USA: Henry T. Lynch, Carrie L. Snyder. (G-FAST) We wish to thank the technical support of Ilse Coene en Brecht Crombez. (HCSC) We acknowledge Alicia Tosar for her technical assistance(HEBCS) HEBCS would like to thank Dr Kristiina Aittomäki, Taru A. Muranen, Drs Carl Blomqvist and Kirsimari Aaltonen and RNs Irja Erkkilä and Virpi Palola for their help with the HEBCS data and samples. (HEBON) The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) consists of the following Collaborating Centres: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: M.A. Rookus, F.B.L. Hogervorst, F.E. van Leeuwen, S. Verhoef, M.K. Schmidt, N.S. Russell, J.L. de Lange, R. Wijnands; Erasmus Medical Center, Rotterdam, NL: J.M. Collée, A.M.W. van den Ouweland, M.J. Hooning, C. Seynaeve, C.H.M. van Deurzen, I.M. Obdeijn; Leiden University Medical Center, NL: C.J. van Asperen, J.T. Wijnen, R.A.E.M. Tollenaar, P. Devilee, T.C.T.E.F. van Cronenburg; Radboud University Nijmegen Medical Center, NL: C.M. Kets, A.R. Mensenkamp; University Medical Center Utrecht, NL: M.G.E.M. Ausems, R.B. van der Luijt, C.C. van der Pol; Amsterdam Medical Center, NL: C.M. Aalfs, T.A.M. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, NL: E.B. Gómez-Garcia, M.J. Blok; University Medical Center Groningen, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits, G.H. de Bock; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen; The Netherlands Comprehensive Cancer Organization (IKNL): S. Siesling, J.Verloop; The Dutch Pathology Registry (PALGA): L.I.H. Overbeek. The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research grant NWO 91109024, the Pink Ribbon grants 110005 and 2014-187.WO76, the BBMRI grant NWO 184.021.007/CP46 and the Transcan grant JTC 2012 Cancer 12-054. HEBON thanks the registration teams of IKNL and PALGA for part of the data collection. (HRBCP) We wish to thank Hong Kong Sanatoriuma and Hospital for their continual support. (HUNBOCS) We wish to thank the Hungarian Breast and Ovarian Cancer Study Group members (Janos Papp, Tibor Vaszko, Aniko Bozsik, Timea Pocza, Judit Franko, Maria Balogh, Gabriella Domokos, Judit Ferenczi, Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary) and the clinicians and patients for their contributions to this study.(HVH) We wish to thank the Oncogenetics Group (VHIO), and the High Risk and Cancer Prevention Unit of the University Hospital Vall d'Hebron.(ICO) We wish to thank the ICO Hereditary Cancer Program team led by Dr Gabriel Capella. (INHERIT) We would like to thank Dr Martine Dumont, Martine Tranchant for sample management and skilful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. J.S. and P.S. were part of the QC and Genotyping coordinating group of iCOGS (BCAC and CIMBA). (IPOBCS) We wish to thank Drs Ana Peixoto, Catarina Santos, Patrícia Rocha and Pedro Pinto for their skilful contribution to the study. (KCONFAB) We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. (MODSQUAD) Modifier Study of Quantitative Effects on Disease (MODSQUAD): we acknowledge ModSQuaD members Csilla Szabo (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA); Lenka Foretova and Eva Machackova (Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute and MF MU, Brno, Czech Republic); and Michal Zikan, Petr Pohlreich and Zdenek Kleibl (Oncogynecologic Center and Department of Biochemistry and Experimental Oncology, First Faculty of Medicine, Charles University, Prague, Czech Republic). (MSKCC) Anne Lincoln, Lauren Jacobs. (NICCC) We wish to thank the NICCC National Familial Cancer Consultation Service team led by Sara Dishon, the lab team led by Dr Flavio Lejbkowicz, and the research field operations team led by Dr Mila Pinchev. (NRG Oncology) We thank the investigators of the Australia New Zealand NRG Oncology group. (OCGN) We wish to thank members and participants in the Ontario Cancer Genetics Network for their contributions to the study. (OSU CCG) Leigha Senter, Kevin Sweet, Caroline Craven, and Michelle O'Conor were instrumental in accrual of study participants, ascertainment of medical records and database management. Samples were processed by the OSU Human Genetics Sample Bank. (SEABASS) We would like to thank Yip Cheng Har, Nur Aishah Mohd Taib, Phuah Sze Yee, Norhashimah Hassan and all the research nurses, research assistants and doctors involved in the MyBrCa Study for assistance in patient recruitment, data collection and sample preparation. In addition, we thank Philip Iau, Sng Jen-Hwei and Sharifah Nor Akmal for contributing samples from the Singapore Breast Cancer Study and the HUKM-HKL Study respectively. The Malaysian Breast Cancer Genetic Study is funded by research grants from the Malaysian Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HIR/MOHE/06) and charitable funding from Cancer Research Initiatives Foundation. (SMC) SMC team wishes to acknowledge the assistance of the Meirav Comprehensice breast cancer center team at the Sheba Medical Center for assistance in this study. (SWE-BRCA) Swedish scientists participating as SWE-BRCA collaborators are: from Lund University and University Hospital: Åke Borg, Håkan Olsson, Helena Jernström, Karin Henriksson, Katja Harbst, Maria Soller, Ulf Kristoffersson; from Gothenburg Sahlgrenska University Hospital: Anna Öfverholm, Margareta Nordling, Per Karlsson, Zakaria Einbeigi; from Stockholm and Karolinska University Hospital: Anna von Wachenfeldt, Annelie Liljegren, Annika Lindblom, Brita Arver, Gisela Barbany Bustinza, Johanna Rantala; from Umeå University Hospital: Beatrice Melin, Christina Edwinsdotter Ardnor, Monica Emanuelsson; from Uppsala University: Hans Ehrencrona, Maritta Hellström Pigg, Richard Rosenquist; from Linköping University Hospital: Marie Stenmark-Askmalm, Sigrun Liedgren(UCHICAGO) We wish to thank Cecilia Zvocec, Qun Niu, physicians, genetic counsellors, research nurses and staff of the Cancer Risk Clinic for their contributions to this resource, and the many families who contribute to our programme. (UCLA) We thank Joyce Seldon MSGC and Lorna Kwan, MPH for assembling the data for this study. (UCSF) We would like to thank Dr Robert Nussbaum and the following genetic counsellors for participant recruitment: Beth Crawford, Kate Loranger, Julie Mak, Nicola Stewart, Robin Lee, Amie Blanco and Peggy Conrad. And thanks to Ms. Salina Chan for her data management. (UKFOCR) We thank Carole Pye, Patricia Harrington and Eva Wozniak for their contributions towards the UKFOCR. (VFCTG) Geoffrey Lindeman, Marion Harris, Martin Delatycki of the Victorian Familial Cancer Trials Group. We thank Sarah Sawyer and Rebecca Driessen for assembling this data and Ella Thompson for performing all DNA amplification.
Grant Support: The COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175—HEALTH-F2-2009-223175). BCAC is funded by Cancer Research UK [C1287/A10118, C1287/A12014] and by the European Community´s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The CIMBA data management and analytical work is funded by Cancer Research UK (C12292/A11174, C12292/A20861). Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding for this project were in part supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium. Funding for the project was provided by the Wellcome Trust under award 076113. The results published here are in part based on data generated by The Cancer Genome Atlas Project established by the National Cancer Institute and National Human Genome Research Institute.
Personal support: K.L. is supported by a K99/R00 grant from the National Cancer Institute (Grant number 1K99CA184415-01). This project was supported in part by a Program Project Development Grant from the Ovarian Cancer Research Fund (S.A.G and A.M). The in vitro aspects of this project were performed within the Norris Cancer Centre at USC, which is supported in part by award number P30CA014089 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. D.F.E. is a Principal Research Fellow of Cancer Research UK. A.C.A. is a Cancer Research—UK Senior Cancer Research Fellow. G.C.-T. and P.M.W. are supported by the National Health and Medical Research Council. (WCP) B.Y.K is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124. L.E.K. is supported by a Canadian Institutes of Health Research Investigator award (MSH-87734). S.P.K. is supported by a Gates Cambridge Scholarship. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. RB was a Cancer Institute NSW Clinical Research Fellow. M.C.S. is a NHMRC Senior Research Fellow. A.K.G. was funded by 5U01CA113916, R01CA140323, and by the Chancellors Distinguished Chair in Biomedical Sciences Professorship. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. S.L.E. and J.D.F. are supported by Fellowships from the National Breast Cancer Foundation (NBCF) Australia and NHMRC project grant (1058415).
Funding: BCAC: The Australian Breast Cancer Family Study (ABCFS) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]. The ACP study is funded by the Breast Cancer Research Trust, UK. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN).ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre.The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev HospitalThe CNIO-BCS was supported by the Instituto de Salud Carlos III, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI12/00070). The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. HAC receives support from the Lon V Smith Foundation (LVS39420). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GC-HBOC was supported by Deutsche Krebshilfe (107 352). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, and by the strategic funding of the University of Eastern Finland. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia, Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; 400413, 400281, 199600). LAABC is supported by grants (1RB-0287, 3PB-0102, 5PB-0018, 10PB-0098) from the California Breast Cancer Research Program. Incident breast cancer cases were collected by the USC Cancer Surveillance Program (CSP), which is supported under subcontract by the California Department of Health. The CSP is also part of the National Cancer Institute's Division of Cancer Prevention and Control Surveillance, Epidemiology, and End Results Program, under contract number N01CN25403. LMBC is supported by the 'Stichting tegen Kanker' (232-2008 and 196-2010). Diether Lambrechts is supported by the FWO and the KULPFV/10/016-SymBioSysII.The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. (MBCSG) is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects '5x1000'). The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the 'CIHR Team in Familial Risks of Breast Cancer' program – grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade – grant # PSR-SIIRI-701.MYBRCA is funded by research grants from the Malaysian Ministry of Science, Technology and Innovation (MOSTI), Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation (CARIF). Additional controls were recruited by the Singapore Eye Research Institute, which was supported by a grant from the Biomedical Research Council (BMRC08/1/35/19/550), Singapore and the National medical Research Council, Singapore (NMRC/CG/SERI/2010). The NBHS was supported by NIH grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland (grant number 250083, 122715 and Center of Excellence grant number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities. The Ontario Familial Breast Cancer Registry (OFBCR) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast CancerThe RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SBCGS was supported primarily by NIH grants R01CA64277, R01CA148667, and R37CA70867. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The scientific development and funding of this project were, in part, supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network U19 CA148065.The SBCS was supported by Yorkshire Cancer Research S295, S299, S305PA and Sheffield Experimental Cancer Medicine Centre.SEARCH is funded by a programme grant from Cancer Research UK [C490/A10124] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge.SGBCC is funded by the NUS start-up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant and the NMRC Clinician Scientist Award. Additional controls were recruited by the Singapore Consortium of Cohort Studies-Multi-ethnic cohort (SCCS-MEC), which was funded by the Biomedical Research Council, grant number: 05/1/21/19/425.SKKDKFZS is supported by the DKFZ. The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation, the Hellenic Cooperative Oncology Group research grant (HR R_BG/04) and the Greek General Secretary for Research and Technology (GSRT) Program, Research Excellence II, the European Union (European Social Fund – ESF), and Greek national funds through the Operational Program 'Education and Lifelong Learning' of the National Strategic Reference Framework (NSRF) - ARISTEIA. The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre.
Funding: OCAC: Funding of the constituent studies was provided by the American Cancer Society (CRTG-00-196-01-CCE); the California Cancer Research Program (00-01389 V-20170, N01-CN25403, 2II0200); the Canadian Institutes for Health Research (MOP-86727); Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124, C536/A13086, C536/A6689); the Celma Mastry Ovarian Cancer Foundation; the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; Imperial Experimental Cancer Research Centre (C1312/A15589); the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L and S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation; the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA15083, P50-CA105009, P50- CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01- CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063682, R01-CA064277, R01-CA067262, R01- CA071766, R01-CA074850, R01-CA076016, R01-CA080742, R01-CA080978, R01-CA083918, R01-CA087538, R01- CA092044, R01-095023, R01-CA106414, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01- CA136924, R01-CA149429, R03-CA113148, R03-CA115195, R37-CA070867, R37-CA70867, U01-CA069417, U01- CA071966, R01-CA063678 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17-98-1- 8659, DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-10-1-0280); the National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01 GB 9401); the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital 'Womens Health Theme' and the Royal Marsden Hospital; WorkSafeBC.
Funding: CIMBA (BCFR—all): This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. (BFBOCC-LT) BFBOCC is partly supported by: Lithuania (BFBOCC-LT): Research Council of Lithuania grant LIG-07/2012; (BIDMC) BIDMC is supported by the Breast Cancer Research Foundation. (BMBSA) BRCA-gene mutations and breast cancer in South African women (BMBSA) was supported by grants from the Cancer Association of South Africa (CANSA) to Elizabeth J. van Rensburg. (BRICOH) SLN was partially supported by the Morris and Horowitz Familes Endowed Professorship. (CBCS) This work was supported by the NEYE Foundation. (CNIO) This work was partially supported by Spanish Association against Cancer (AECC08), RTICC 06/0020/1060, FISPI08/1120, Mutua Madrileña Foundation (FMMA) and SAF2010-20493(COH-CCGCRN) City of Hope Clinical Cancer Genetics Community Network and the Hereditary Cancer Research Registry, supported in part by Award Number RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. (CONSIT TEAM) Funds from Italian citizens who allocated the 5x1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects '5x1000') to Siranoush Manoukian. (CORE) The CIMBA data management and data analysis were supported by Cancer Research – UK grants C12292/A11174 and C1287/A10118.SH is supported by an NHMRC Program Grant to GCT. ACA is a Cancer Research -UK Senior Cancer Research Fellow. (DEMOKRITOS) This research has been co-financed by the European Union (European Social Fund – ESF) and Greek national funds through the Operational Program 'Education and Lifelong Learning' of the National Strategic Reference Framework (NSRF) - Research Funding Program of the General Secretariat for Research and Technology: ARISTEIA. Investing in knowledge society through the European Social Fund.(DKFZ) The DKFZ study was supported by the DKFZ. (EMBRACE) EMBRACE is supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. (FCCC) The authors acknowledge support from The University of Kansas Cancer Center (P30 CA168524) and the Kansas Bioscience Authority Eminent Scholar Program. (GC-HBOC) The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC) is supported by the German Cancer Aid (grant no 109076, Rita K. Schmutzler) and by the Center for Molecular Medicine Cologne (CMMC). (GEMO) The study was supported by the Ligue Nationale Contre le Cancer; the Association 'Le cancer du sein, parlons-en!' Award; the Canadian Institutes of Health Research for the 'CIHR Team in Familial Risks of Breast Cancer' program and the French National Institute of Cancer (INCa). (GEORGETOWN) CI received support from the Non-Therapeutic Subject Registry Shared Resource at Georgetown University (NIH/NCI grant P30-CA051008), the Fisher Center for Familial Cancer Research, and Swing Fore the Cure. (G-FAST) Kim De Leeneer is supported by GOA grant BOF10/GOA/019 (Ghent University) and spearhead financing of Ghent University Hospital. (HCSC) HCSC supported by a grant RD12/0036/0006 and 12/00539 from ISCIII (Spain), partially supported by European Regional Development FEDER funds. (HEBCS) The HEBCS was financially supported by the Helsinki University Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society and the Sigrid Juselius Foundation. (HEBON) The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research grant NWO 91109024, the Pink Ribbon grant 110005 and the BBMRI grant NWO 184.021.007/CP46. HEBON thanks the registration teams of the Comprehensive Cancer Centre Netherlands and Comprehensive Centre South (together the Netherlands Cancer Registry) and PALGA (Dutch Pathology Registry) for part of the data collection. (HRBCP) HRBCP is supported by The Hong Kong Hereditary Breast Cancer Family Registry and the Dr Ellen Li Charitable Foundation, Hong Kong (HUNBOCS) Hungarian Breast and Ovarian Cancer Study was supported by Hungarian Research Grants KTIA-OTKA CK-80745, OTKA K-112228 and the Norwegian EEA Financial Mechanism Hu0115/NA/2008-3/OP-9. (ICO) Contract grant sponsor: Asociación Española Contra el Cáncer, Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia. Contract grant numbers: ISCIIIRETIC RD06/0020/1051, RD12/0036/008, PI10/01422, PI10/00748, PI13/00285, PIE13/00022, 2009SGR290 and 2014SGR364. (IHCC) The IHCC was supported by Grant PBZ_KBN_122/P05/2004(ILUH) The ILUH group was supported by the Icelandic Association 'Walking for Breast Cancer Research' and by the Landspitali University Hospital Research Fund. (INHERIT) This work was supported by the Canadian Institutes of Health Research for the 'CIHR Team in Familial Risks of Breast Cancer' program, the Canadian Breast Cancer Research Alliance-grant #019511 and the Ministry of Economic Development, Innovation and Export Trade – grant # PSR-SIIRI-701. (IOVHBOCS) IOVHBOCS is supported by Ministero della Salute and '5 × 1,000' Istituto Oncologico Veneto grant. (IPOBCS) This study was in part supported by Liga Portuguesa Contra o Cancro.(KCONFAB) kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia; (KOHBRA) KOHBRA is supported by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (1020350). (MAYO) MAYO is supported by NIH grants CA116167, CA128978 and CA176785, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a U.S. Department of Defence Ovarian Cancer Idea award (W81XWH-10-1-0341), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. (MCGILL) Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade (MODSQUAD) MODSQUAD was supported by MH CZ - DRO (MMCI, 00209805) and by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101) to LF, and by Charles University in Prague project UNCE204024 (MZ). (MSKCC) MSKCC is supported by grants from the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Cancer Genetics Initiative, and the Andrew Sabin Research Fund. (NAROD) 1R01 CA149429-01. (NCI) The research of Drs MH Greene, JT Loud and PL Mai was supported by the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts NO2-CP-11019-50 and N02-CP-65504 with Westat, Inc, Rockville, MD. (NICCC) NICCC is supported by Clalit Health Services in Israel. Some of it's activities are supported by the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), NY. (NNPIO) This work has been supported by the Russian Federation for Basic Research (grants 13-04-92613, 14-04-93959 and 15-04-01744). (NRG Oncology) This study was supported by National Cancer Institute grants to the NRG Oncology Administrative Office and Tissue Bank (CA 27469), the NRG Oncology Statistical and Data Center (CA 37517), and NRG Oncology's Cancer Prevention and Control Committee (CA 101165). (OSU CCG) OSUCCG is supported by the Ohio State University Comprehensive Cancer Center. (PBCS) This work was supported by the ITT (Istituto Toscano Tumori) grants 2011-2013. (SEABASS) Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation. (SMC) This project was partially funded through a grant by the Isreal cancer association and the funding for the Israeli Inherited breast cancer consortium (SWE-BRCA) SWE-BRCA collaborators are supported by the Swedish Cancer Society. (UCHICAGO) UCHICAGO is supported by NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women's Cancer Research Alliance and the Breast Cancer research Foundation. OIO is an ACS Clinical Research Professor.(UCLA) Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. (UCSF) UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. (UKFOCR) UKFOCR was supported by a project grant from CRUK to Paul Pharoah. (UPENN) National Institutes of Health (NIH) (R01-CA102776 and R01-CA083855; Breast Cancer Research Foundation; Susan G. Komen Foundation for the cure, Basser Research Center for BRCA. (UPITT/MWH) Frieda G. and Saul F. Shapira BRCA-Associated Cancer Research Program;Hackers for Hope Pittsburgh. (VFCTG) Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund and grants R01-CA122443 and P50-CA136393.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-4, Supplementary Tables 1-10 and Supplementary References. (PDF 1359 kb)
Supplementary Data 1
EQTL analyses in normal breast tissues from the METABRIC study. P-values from linear regression analyses. (XLSX 49 kb)
Supplementary Data 2
Expression QTL analyses in normal primary ovarian and fallopian tube secretory epithelial cells, using Fluidigm gene expression chips and TaqMan probes. Positive results were verified using the 7900HT fast real-time PCR system (Life Technologies). # The positive result for MYO9B was not reproducible but † the ANKLE1 association was verified (P=0.002). (XLSX 41 kb)
Supplementary Data 3
Significant allele specific expression in breast cancer samples from TCGA (Peak 1) (P<0.05, F-tests). No associations were observed for any Peak 2 SNPs. (XLSX 38 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Reprints and Permissions
About this article
Cite this article
Lawrenson, K., Kar, S., McCue, K. et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus. Nat Commun 7, 12675 (2016). https://doi.org/10.1038/ncomms12675
Download citation
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/ncomms12675
Further reading
-
Cytogenomic characteristics of murine breast cancer cell line JC
- Shaymaa Azawi
- Martina Rincic
- Thomas Liehr
Molecular Cytogenetics (2021)
-
A search for modifying genetic factors in CHEK2:c.1100delC breast cancer patients
- Camilla Wendt
- Taru A. Muranen
- Annika Lindblom
Scientific Reports (2021)
-
NGS zur Selektion innovativer Therapien – Was bringt das?
- Carla E. Schulmeyer
- Simon Bader
- Peter A. Fasching
Der Gynäkologe (2021)
-
Allele-specific miRNA-binding analysis identifies candidate target genes for breast cancer risk
- Ana Jacinta-Fernandes
- Joana M. Xavier
- Ana-Teresa Maia
npj Genomic Medicine (2020)
-
Functional Landscape of Common Variants Associated with Susceptibility to Epithelial Ovarian Cancer
- Paulo Cilas Morais Lyra
- Letícia B. Rangel
- Alvaro N. A. Monteiro
Current Epidemiology Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Boutry Hofmann Norris the West Guide to Writing Pdf
Source: https://www.nature.com/articles/ncomms12675?origin=ppub
0 Response to "Boutry Hofmann Norris the West Guide to Writing Pdf"
Postar um comentário